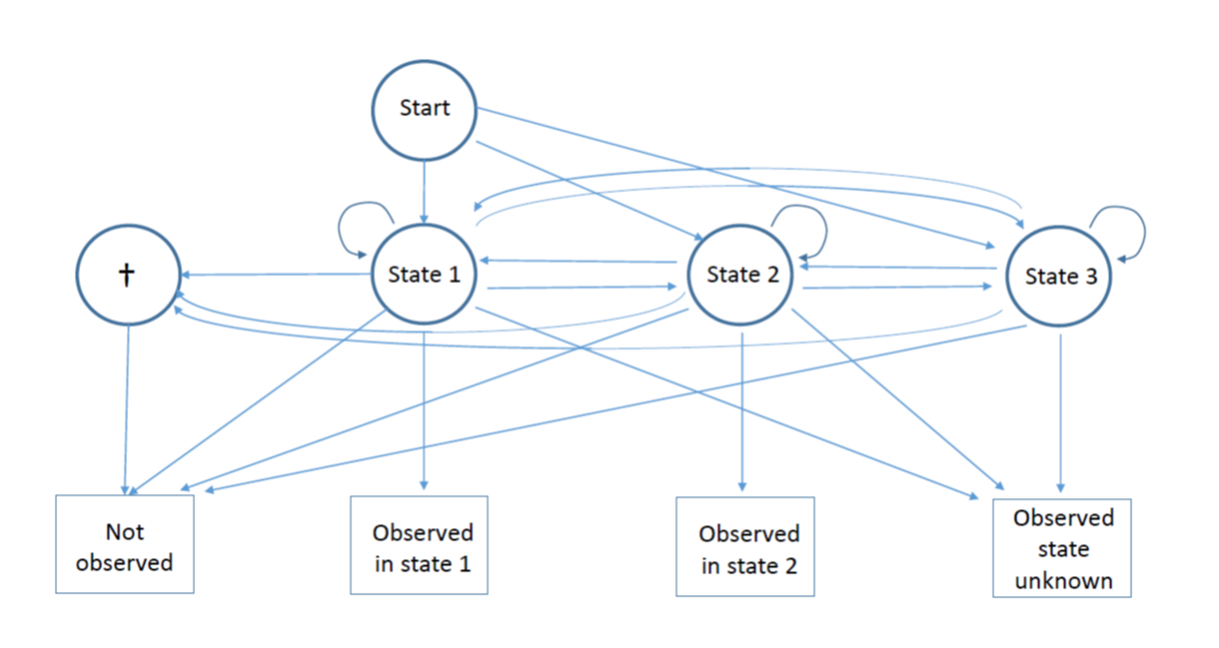
Former SE@K PhD student Anita Jeyam, Rachel and Roger Pradel (Montpellier) have had a paper published presenting a new test for determining the underlying state structure of Hidden Markov models. This is an exciting piece of work which provides the foundation for addressing a complex topic for general HMMs.
The full paper is available open access here: https://www.frontiersin.org/articles/10.3389/fevo.2021.598325/full
A Test for the Underlying State-Structure of Hideen Markov Models – Partially Observed Capture-Recapture Data
Hidden Markov models (HMMs) are being widely used in the field of ecological modeling,
however determining the number of underlying states in an HMM remains a challenge.
Here we examine a special case of capture-recapture models for open populations,
where some animals are observed but it is not possible to ascertain their state (partial
observations), whilst the other animals’ states are assigned without error (complete
observations). We propose a mixture test of the underlying state structure generating
the partial observations, which assesses whether they are compatible with the set of
states observed in the complete observations. We demonstrate the good performance
of the test using simulation and through application to a data set of Canada Geese.