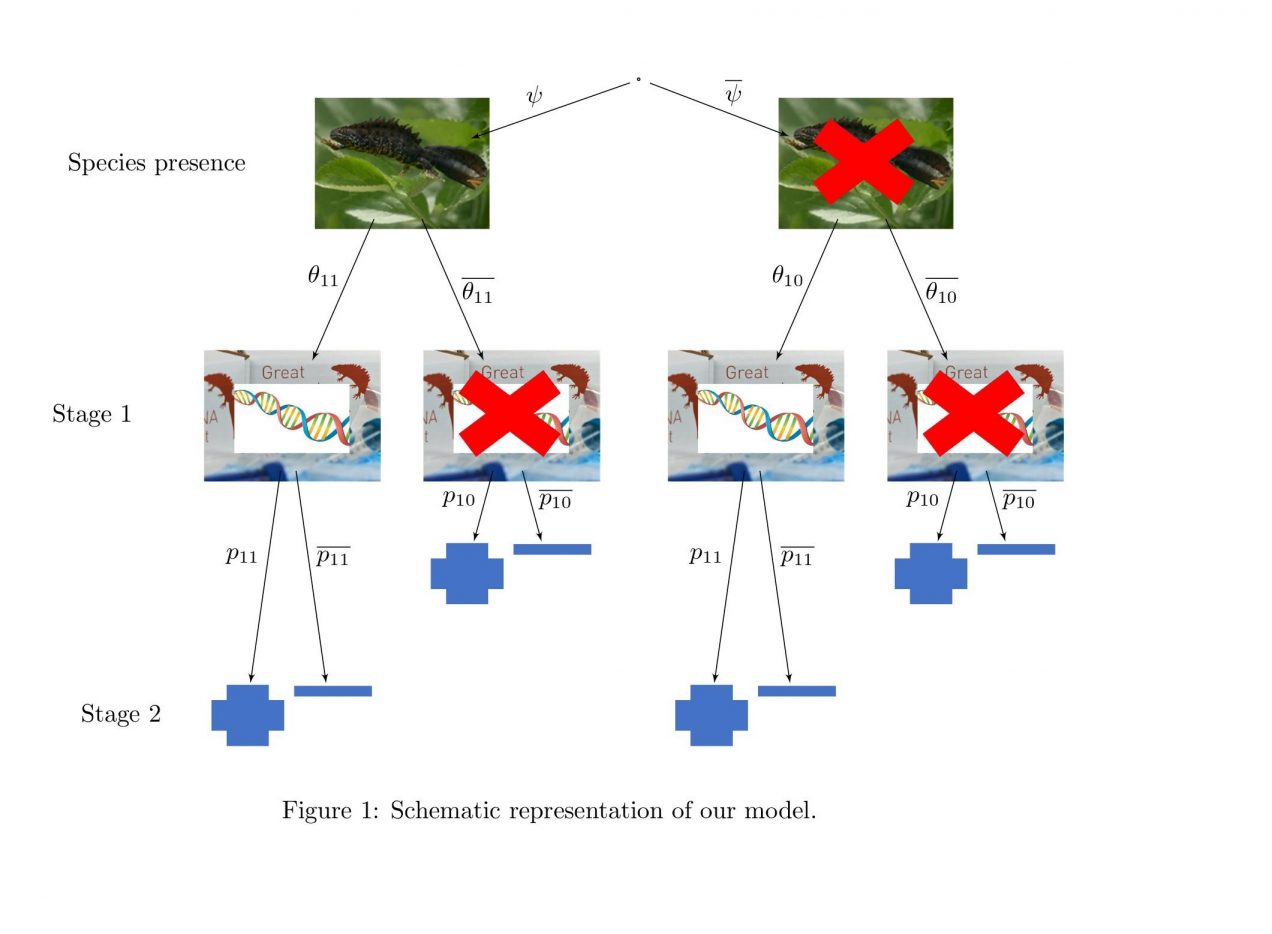
Environmental DNA (eDNA) is an increasingly popular survey tool for monitoring species distribution. eDNA surveys have been used with a wide variety of species in different landscapes and there is growing evidence that they suffer from lower observation error than existing methods relying on direct observation of the target species.
From detecting single species using quantitative polymerase chain reaction (qPCR), to studying whole communities using metabarcoding, eDNA is showing great promise in helping us understand species distributions and community compositions.
However, we are yet to fully understand the properties of eDNA, and hence are only beginning to appreciate the opportunities that eDNA surveys bring or the challenges that we need to overcome in the field, in the lab or in implementing eDNA surveys into policy.
This meeting brings together researchers who are leading in the development of new statistical methods for analysing eDNA data, in evaluating the use of eDNA surveys with different species and landscapes, or in embedding eDNA techniques into national or international policy.
Speakers and talks
- 10.15-11 Kerry Walsh, Environment Agency: “Challenges and opportunities: A regulator’s perspective.”
- 11-11.30 break and refreshments
- 11.30-12.15 Naomi Ewald, FreshWater Habitats Trust: “Analysis of eDNA data to inform conservation priorities: case studies of long term species monitoring and short term before-after surveys.”
- 12.15-13 Francesco Ficetola, University of Milan: “Environmental DNA to track long-term changes of mountain ecosystem.”
- 13-14 lunch
- 14-14.45 Jim Griffin, University College London: “Modelling environmental DNA data; Bayesian variable selection accounting for false positive and false negative errors.”
- 14.45-15.30
- Doug Yu, University of East Anglia: “Managing wildlife with eDNA data: salmon, leeches, insects, and forests.”
- 15.30-16.00 Discussion
The meeting, organised by the Environmental Statistics Section and the Emerging Applications Section of the Royal Statistical Society (RSS) will take place on the 7th of May 2020 at the RSS headquarters (12 Errol St, London EC1Y 8LX).
Follow this link to register for the event.
If you have any questions email e.matechou@kent.ac.uk.