The workshop, in Montpellier, France, was organised by Matt Silk and Olivier Gimenez and was attended by around 12 experts in statistical models for ecological data and in network modelling. 


Category Archives: Uncategorized
rGAI: An R package for fitting the generalized abundance index to seasonal count data
Emily Dennis, Calliste Fagard-Jenkin and Byron Morgan have created an R package for fitting the generalized abundance index to seasonal count data. The work has been published in Ecology and Evolution in the paper “rGAI: An R package for fitting the generalized abundance index to seasonal count data”.
The paper can be found at: https://onlinelibrary.wiley.com/doi/full/10.1002/ece3.9200
The R package is available at: https://github.com/calliste-fagard-jenkin/rGAI
Statistical Ecology Conference
Members of SE@K group attended ISEC from 27th June to 1st July. Some went to South Africa to attend in person others attended virtually.
Eleni, Alex and Ioannis gave a half day workshop on Modelling environmental DNA data.
Eleni presented work on Capture recapture models with heterogeneous temporary.
Alex gave a talk on A unifying modelling framework for metabarcoding data.
Rachel talked on Model selection for integrated population models: selecting age structure with multiple data types.
James’ presentation was on Accounting for varying spatial scales in the production of UK butterfly abundance estimates.
Diana talked about Bayesian Identifiability in Ecological Models.
Fay presented work on Assessing the success of reintroductions whilst accounting for multispecies populations.
Bryon talked about Fitting dynamic occupancy models to very large occurrence data sets using hidden Markov models.

Obituaries for Philip North
Philip North, 21st May, 1949 – 4th June, 2021
It is our sad duty to let you know of the death of Philip North, who died on the 4th June 2021, aged 72. Philip made a huge contribution to the development of the EURING Analytical Meetings, acting as editor of the first five proceedings.
He obtained his PhD on Statistical methods in ornithology from the University of Kent in 1979. This involved the development of novel analyses of various BTO datasets, in particular devising a method of cluster analysis to determine bird territories and enhancing various aspects of survival estimation. His influential 1978 paper in Biometrics used a variety of methods, including logistic regression and point process modelling, to investigate the effects of weather on the survival of grey herons. In addition, through his enthusiasm and research highly productive links were established between the Universities of Kent and St Andrews and with the Centre d’Ecologie Fontionnelle & Evolutive, CNRS Montpellier, which, together with the BTO are continued by the current members of the Statistical Ecology at Kent research group.
Philip was an expert ornithologist and a keen birder. For example, we recall that he was one of a small number of observers who recorded the first sighting of a pallid swift in Britain and Ireland Thus he was well placed to understand both the statistical and ecological aspects of avian ecology. This combination of ornithology and statistical modeling made him an ideal contributor to the EURING Analytical Conferences, as did his role as secretary of the Mathematical Ecology Group of the British Region of the Biometric Society and the British Ecological Society.
Innovative discussions and published proceedings became key features of these EURING meetings. The first two were held in Wageningen in 1986 and at Sempach in 1989 and Philip edited the resulting proceedings single-handed, as well as jointly editing the proceedings of the meetings held in Montpellier (1992), Patuxent (1994) and Norwich (1997; see https://euring.org/meetings/analytical-meetings/analytical-meeting-proceedings ) . This was an important and sustained contribution, which appreciably advanced the theory and application of relevant aspects of statistical ecology, with wide application to both theoretical and applied problems. It also showed how data gathered by the bird ringing/banding schemes coordinated through EURING could be collected and analyzed in ways that enhanced their ecological value. It is particularly noticeable how the content of the conference papers evolved over Philip’s period as editor. The first meeting focused on the use of individual datasets to estimate specific parameters, mainly survival. By the 1997 meeting there was greater interest in model selection and applied studies, with the first signs of the developments in data integration that would follow over the next 20 years.
Philip’s wife Monica died in 2008. They are survived by their children, Robin, Geoffrey and Melissa to whom we extend our deep condolences.
An obituary has recently appeared in JRSSA https://rss.onlinelibrary.wiley.com/doi/10.1111/rssa.12767
Byron Morgan and Stephen Baillie
Think Piece with Eleni as co-author published on Gov.uk
The Think Piece, titled “Understanding ecosystems and resilience using DNA“, explores opportunities for applying advances in DNA and RNA technologies to improve understanding of ecosystem function and resilience. Eleni’s contribution in particular, written together with Doug Yu, UEA, looks at “The contribution of DNA-based methods to achieving socio-ecological resilience”.
New Paper: A guide to state–space modeling of ecological time series
Diana is co-author on the paper: A guide to state–space modeling of ecological time series, which was published in Ecological Monographs.
https://esajournals.onlinelibrary.wiley.com/doi/full/10.1002/ecm.1470
Abstract: State–space models (SSMs) are an important modeling framework for analyzing ecological time series. These hierarchical models are commonly used to model population dynamics, animal movement, and capture–recapture data, and are now increasingly being used to model other ecological processes. SSMs are popular because they are flexible and they model the natural variation in ecological processes separately from observation error. Their flexibility allows ecologists to model continuous, count, binary, and categorical data with linear or nonlinear processes that evolve in discrete or continuous time. Modeling the two sources of stochasticity separately allows researchers to differentiate between biological variation and imprecision in the sampling methodology, and generally provides better estimates of the ecological quantities of interest than if only one source of stochasticity is directly modeled. Since the introduction of SSMs, a broad range of fitting procedures have been proposed. However, the variety and complexity of these procedures can limit the ability of ecologists to formulate and fit their own SSMs. We provide the knowledge for ecologists to create SSMs that are robust to common, and often hidden, estimation problems, and the model selection and validation tools that can help them assess how well their models fit their data. We present a review of SSMs that will provide a strong foundation to ecologists interested in learning about SSMs, introduce new tools to veteran SSM users, and highlight promising research directions for statisticians interested in ecological applications. The review is accompanied by an in-depth tutorial that demonstrates how SSMs can be fitted and validated in R. Together, the review and tutorial present an introduction to SSMs that will help ecologists to formulate, fit, and validate their models.
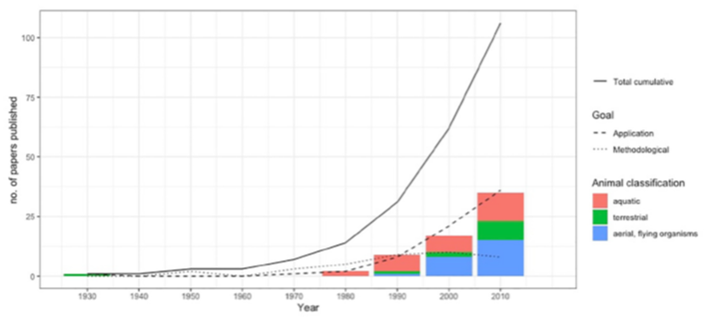
New paper: Removal modelling in ecology – A systematic review
SE@K members Oscar and Rachel have published a paper reviewing the current state-of-the-art in modelling removal data. This work is part of the the EPSRC project EP/S020470/1 “Modelling removal and re-introduction data for improved conservation”.
The paper is available in full here: https://journals.plos.org/plosone/article/authors?id=10.1371/journal.pone.0229965
Removal modelling in ecology: A systematic review
Oscar Rodriguez de Rivera and Rachel McCrea
Removal models were proposed over 80 years ago as a tool to estimate unknown population size. More recently, they are used as an effective tool for management actions for the control of non desirable species, or for the evaluation of translocation management actions. Although the models have evolved over time, in essence, the protocol for data collection has remained similar: at each sampling occasion attempts are made to capture and remove individuals from the study area. Within this paper we review the literature of removal modelling and highlight the methodological developments for the analysis of removal data, in order to provide a unified resource for ecologists wishing to implement these approaches. Models for removal data have developed to better accommodate important features of the data and we discuss the shift in the required assumptions for the implementation of the models. The relative simplicity of this type of data and associated models mean that the method remains attractive and we discuss the potential future role of this technique.
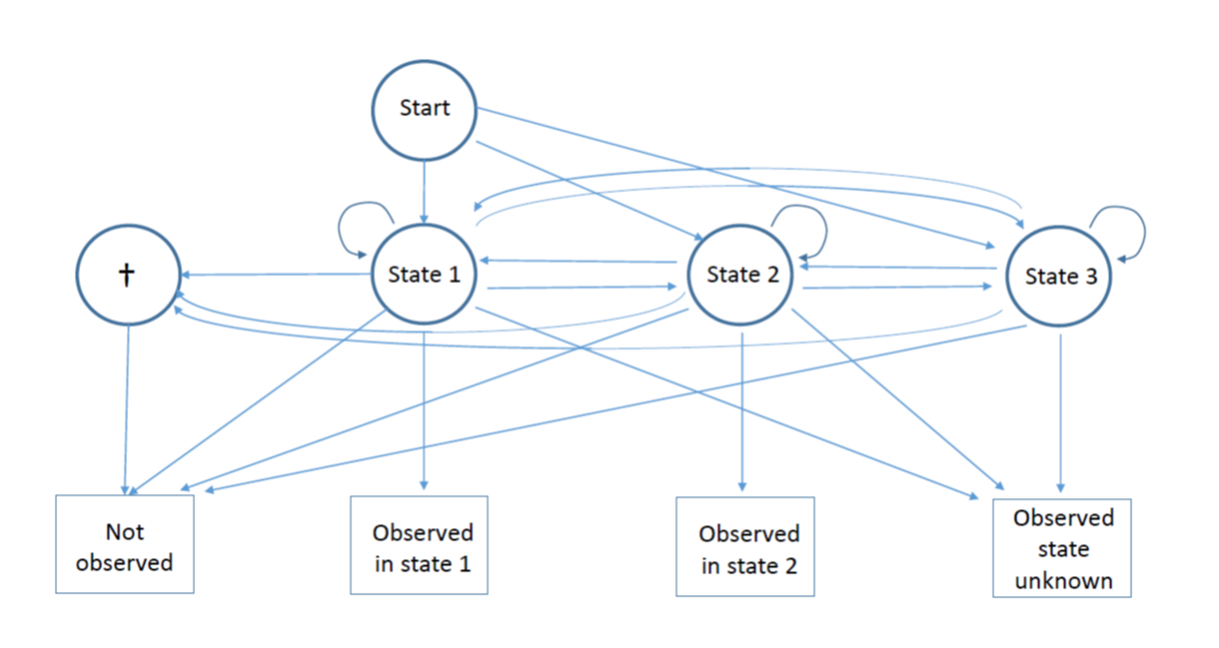
New paper: A Test for the Underlying State-Structure of Hideen Markov Models – Partially Observed Capture-Recapture Data
Former SE@K PhD student Anita Jeyam, Rachel and Roger Pradel (Montpellier) have had a paper published presenting a new test for determining the underlying state structure of Hidden Markov models. This is an exciting piece of work which provides the foundation for addressing a complex topic for general HMMs.
The full paper is available open access here: https://www.frontiersin.org/articles/10.3389/fevo.2021.598325/full
A Test for the Underlying State-Structure of Hideen Markov Models – Partially Observed Capture-Recapture Data
Hidden Markov models (HMMs) are being widely used in the field of ecological modeling,
however determining the number of underlying states in an HMM remains a challenge.
Here we examine a special case of capture-recapture models for open populations,
where some animals are observed but it is not possible to ascertain their state (partial
observations), whilst the other animals’ states are assigned without error (complete
observations). We propose a mixture test of the underlying state structure generating
the partial observations, which assesses whether they are compatible with the set of
states observed in the complete observations. We demonstrate the good performance
of the test using simulation and through application to a data set of Canada Geese.

New paper: Modeling Recruitment of Birth Cohorts to the Breeding Population: A Hidden Markov Approach
Rachel McCrea and co-authors from St Andrews and Edinburgh have published a paper on modelling the recruitment process of gray seals.
The full paper is available open access here: https://www.frontiersin.org/articles/10.3389/fevo.2021.600967/full
Modeling Recruitment of Birth Cohorts to the Breeding Population: A Hidden Markov Approach
Worthington, King, McCrea, Smout and Pomeroy
Long-term capture-recapture studies provide an opportunity to investigate the population dynamics of long-lived species through individual maturation and adulthood and/or
time. We consider capture-recapture data collected on cohorts of female gray seals
(Halichoerus grypus) born during the 1990s and later observed breeding on the Isle
of May, Firth of Forth, Scotland. Female gray seals can live for 30+ years but display
individual variability in their maturation rates and so recruit into the breeding population
across a range of ages. Understanding the partially hidden process by which individuals
transition from immature to breeding members, and in particular the identification of
any changes to this process through time, are important for understanding the factors
affecting the population dynamics of this species. Age-structured capture-recapture
models can explicitly relate recruitment, and other demographic parameters of interest,
to the age of individuals and/or time. To account for the monitoring of the seals from
several birth cohorts we consider an age-structured model that incorporates a specific
cohort-structure. Within this model we focus on the estimation of the distribution of
the age of recruitment to the breeding population at this colony. Understanding this
recruitment process, and identifying any changes or trends in this process, will offer
insight into individual year effects and give a more realistic recruitment profile for the
current UK gray seal population model. The use of the hidden Markov model provides an
intuitive framework following the evolution of the true underlying states of the individuals.
The model breaks down the different processes of the system: recruitment into the
breeding population; survival; and the associated observation process. This model
specification results in an explicit and compact expression for the model with associated
efficiency in model fitting. Further, this framework naturally leads to extensions to more
complex models, for example the separation of first-time from return breeders, through
relatively simple changes to the mathematical structure of the model.
New Paper: Parameter redundancy in Jolly‐Seber tag loss models
Diana along with Wei Cai, Stephanie Yurchak and Laura Cowen have published the paper:
Parameter redundancy in Jolly‐Seber tag loss models
in Ecology and Evolution
Abstract:
1. Capture–recapture experiments are conducted to estimate population parameters such as population size, survival rates, and capture rates. Typically, individuals are captured and given unique tags, then recaptured over several time periods with the assumption that these tags are not lost. However, for some populations, tag loss cannot be assumed negligible. The Jolly‐Seber tag loss model is used when the no‐tag‐loss assumption is invalid. Further, the model has been extended to incorporate group heterogeneity, which allows parameters to vary by group membership. Many mark–recapture models become overparameterized resulting in the inability to independently estimate parameters. This is known as parameter redundancy.
2. We investigate parameter redundancy using symbolic methods. Because of the complex structure of some tag loss models, the methods cannot always be applied directly. Instead, we develop a simple combination of parameters that can be used to investigate parameter redundancy in tag loss models.
3. The incorporation of tag loss and group heterogeneity into Jolly‐Seber models does not result in further parameter redundancies. Furthermore, using hybrid methods we studied the parameter redundancy caused by data through case studies and generated tag histories with different parameter values.
4. Smaller capture and survival rates are found to cause parameter redundancy in these models. These problems resolve when applied to large populations.
