The final report of the landscape decisions programme, which funded Eleni’s NERC project
Category Archives: News

Metabarcoding paper published in JASA
eDNAPlus: A Unifying Modeling Framework for DNA-based Biodiversity Monitoring
by Alex Diana, Eleni Matechou, Jim Griffin, Doug Yu, Richard Griffiths and others.
Abstract
DNA-based biodiversity surveys, which involve collecting physical samples from survey sites and assaying them in the laboratory to detect species via their diagnostic DNA sequences, are increasingly being adopted for biodiversity monitoring and decision-making. The most commonly employed method, metabarcoding, combines PCR with high-throughput DNA sequencing to amplify and read “DNA barcode” sequences, generating count data indicating the number of times each DNA barcode was read. However, DNA-based data are noisy and error-prone, with several sources of variation, and cannot alone estimate the species-specific amount of DNA present at a surveyed site (DNA biomass). In this article, we present a unifying modeling framework for DNA-based survey data that allows estimation of changes in DNA biomass within species, across sites and their links to environmental covariates, while for the first time simultaneously accounting for key sources of variation, error and noise in the data-generating process, and for between-species and between-sites correlation. Bayesian inference is performed using MCMC with Laplace approximations. We describe a re-parameterization scheme for crossed-effects models designed to improve mixing, and an adaptive approach for updating latent variables, which reduces computation time. Theoretical and simulation results are used to guide study design, including the level of replication at different survey stages and the use of quality control methods. Finally, we demonstrate our new framework on a dataset of Malaise-trap samples, quantifying the effects of elevation and distance-to-road on each species, and produce maps identifying areas of high biodiversity and species DNA biomass. Supplementary materials for this article are available online, including a standardized description of the materials available for reproducing the work.

RSS Discussion paper – Analysis of citizen science data
A paper by Emily, Alex, Eleni and Byron was discussed at the 2024 RSS conference in Brighton as part of the multi-paper discussion meeting Analysis of citizen science data.
The meeting was chaired by the president of the RSS, Dr Andrew Garrett.
Alex and Eleni presented the paper, which was discussed by Dr Ben Swallow, St Andrews university, and Professor Kerrie Mengersen, University of Queensland.
Details of the paper are given below
Paper 1: ‘Efficient statistical inference methods for assessing changes in species‘
Authors: Emily B Dennis12, Alex Diana3, Eleni Matechou2, Byron J T Morgan2
(1Butterfly Conservation, 2University of Kent, 3University of Essex)
Download the preprint
Supplementary materials
Abstract: The global decline of biodiversity, driven by habitat degradation and climate breakdown, is a significant concern. Accurate measures of change are crucial to provide reliable evidence of species’ population changes. Meanwhile citizen science data have witnessed a remarkable expansion in both quantity and sources and serve as the foundation for assessing species’ status. The growing data reservoir presents opportunities for novel and improved inference but often comes with computational costs: computational efficiency is paramount, especially as regular analysis updates are necessary. Building upon recent research, we present illustrations of computationally efficient methods for fitting new models, applied to three major citizen science data sets for butterflies. We extend a method for modelling abundance changes of seasonal organisms, firstly to accommodate multiple years of count data efficiently, and secondly for application to counts from a snapshot mass-participation survey. We also present a variational inference approach for fitting occupancy models efficiently to opportunistic citizen science data. The continuous growth of citizen science data offers unprecedented opportunities to enhance our understanding of how species respond to anthropogenic pressures. Efficient techniques in fitting new models are vital for accurately assessing species’ status, supporting policy-making, setting measurable targets, and enabling effective conservation efforts.
Speakers, organisers and discussants enjoyed dinner and interesting conversation at a nearby restaurant.
The session was recorded and is available on the RSS youtube channel (Discussion meetings)
Migration & Movement Signature Research Theme Documentary
Watch the short documentary highlighting the work of the SRT, which also includes members of SE@K, such as Bruno, Daniel and Eleni.

SE@K Thursday lunches – Summer 2023
For the last term of the 22/23 academic year, we have an exciting plan for SE@K and guest talks
- 11th of May Session : eDNA – Alex/Eleni; Cake : Eleni
- 18th of May Session : Occupancy data – Ardian Ardiantiono; Cake : James
- 25th of May Session : BeeWalk modelling – Fabian; Cake : Fabian
- 1st of June Session : Elephant killings – Kennedy Leneuiyia; Cake : Milly
- 8th of June Session : Extinction Models – Dave Roberts; Cake : Bruno
- 15th of June Session : Binary time series – Takis (TBC); Cake : Daniel
- 22nd of June Session : PhD student NCSE presentations; Cake : Diana

Bruno and Eleni visited Stockholm
The visit was part of their Swedish research council funded project on modelling overcoverage in population registers, in collaboration with Dr Eleonora Mussino.
The first paper from this project is already under review, using multiple systems estimation to estimate the number of immigrants who are present but have not been observed in any register each year.
They are currently developing capture-recapture-recovery models with temporary emigration as an alternative approach for estimating the probability that an individual is there given that they weren’t observed.
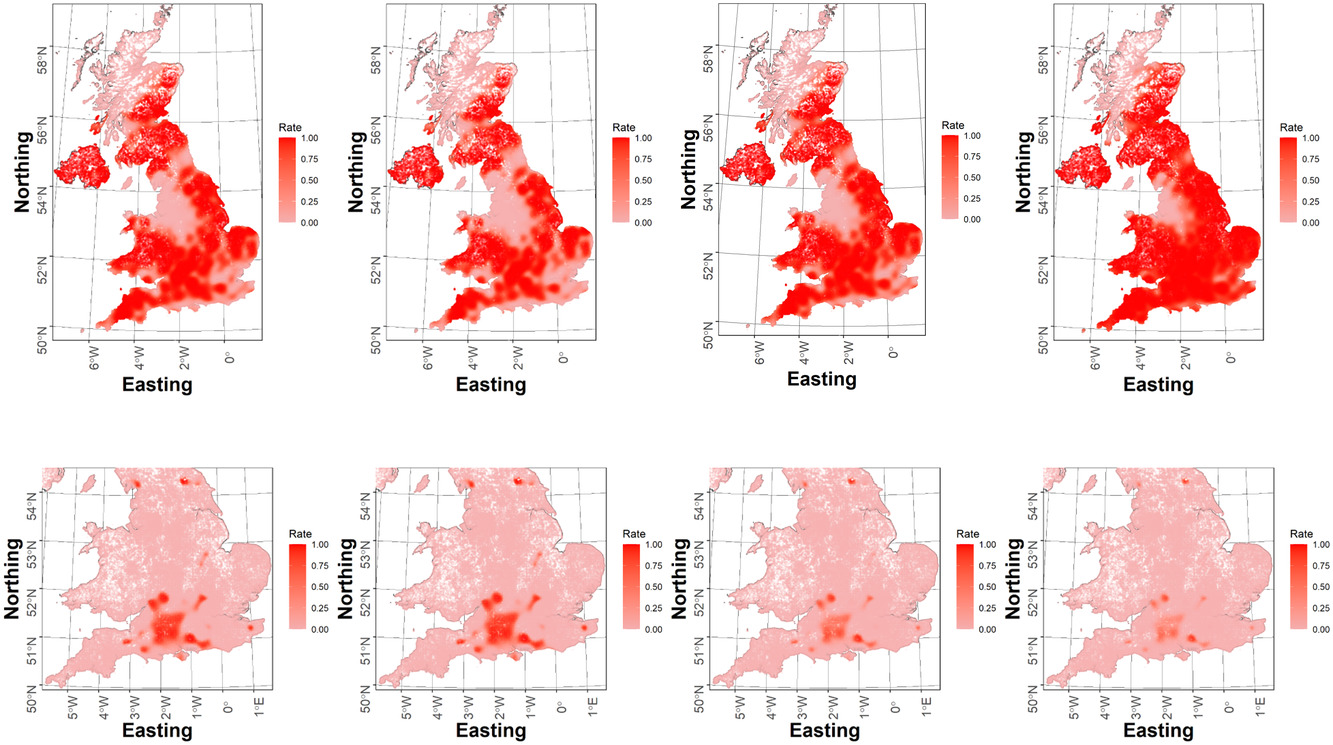
New Biometrics paper
A new modelling framework for fast Bayesian inference in large occupancy data sets by Alex, Emily, Eleni and Byron just published in Biometrics.
The model accounts for spatio-temporal autocorrelation and gives robust inference on species presence, as evidence by the simulation results in the paper and the two case studies on one common and widespread and one rare species, using records from the Butterflies for the New Millennium database, producing occupancy indices spanning 45 years.

Eleni visited Lille for 3i workshop on Biodiversity and its Conservation
The one-day workshop, organised by the group in Lille, consisted of a series of talks by researchers from the Universities of Lille, Ghent and Kent.
Eleni gave talks on her work on modelling BeeWalk data and on modelling eDNA data.

New paper in Environmetrics by Alex
A vector of point processes for modeling interactions between and within species using capture-recapture data
Alex Diana, Eleni Matechou, Jim E. Griffin, Yadvendradev Jhala, Qamar Qureshi
Abstract
Capture-recapture (CR) data and corresponding models have been used extensively to estimate the size of wildlife populations when detection probability is less than 1. When the locations of traps or cameras used to capture or detect individuals are known, spatially-explicit CR models are used to infer the spatial pattern of the individual locations and population density. Individual locations, referred to as activity centers (ACs), are defined as the locations around which the individuals move. These ACs are typically assumed to be independent, and their spatial pattern is modeled using homogeneous Poisson processes. However, this assumption is often unrealistic, since individuals can interact with each other, either within a species or between different species. In this article, we consider a vector of point processes from the general class of interaction point processes and develop a model for CR data that can account for interactions, in particular repulsions, between and within multiple species. Interaction point processes present a challenge from an inferential perspective because of the intractability of the normalizing constant of the likelihood function, and hence standard Markov chain Monte Carlo procedures to perform Bayesian inference cannot be applied. Therefore, we adopt an inference procedure based on the Monte Carlo Metropolis Hastings algorithm, which scales well when modeling more than one species. Finally, we adopt an inference method for jointly sampling the latent ACs and the population size based on birth and death processes. This approach also allows us to adaptively tune the proposal distribution of new points, which leads to better mixing especially in the case of non-uniformly distributed traps. We apply the model to a CR data-set on leopards and tigers collected at the Corbett Tiger Reserve in India. Our findings suggest that between species repulsion is stronger than within species, while tiger population density is higher than leopard population density at the park.