Congratulations to Rachel for being awarded an EPSRC New Investigator award for her project “Modelling removal and re-introduction data for improved conservation”.

Congratulations to Rachel for being awarded an EPSRC New Investigator award for her project “Modelling removal and re-introduction data for improved conservation”.
Click here to submit your application.
Project description: Capture-recapture data are commonly collected and modelled for the study of wildlife populations (McCrea and Morgan, 2014). Existing capture-recapture models typically assume independence between individuals, with no network structure. Hence, the probability of capture, survival, or movement of particular individuals is assumed to be unaffected by events occurring to other individuals, or actions of these individuals, in the same population. The assumption of independence between individuals gives rise to likelihood functions that are simple products of probabilities of individual capture histories.
However, animal populations are structured by interaction networks among individuals and many species tend to live in social groups that result in these interaction networks differing from random (Wey et al., 2008; Pinter-Wollman et al., 2014; Krause et al., 2015;). In these cases, probabilities of certain events involving particular individuals might be related to their position in the social network. For example, individuals that are strongly connected in a network may be more likely to be captured at the same event. However, network relationships may also result in covariation in survival, or associated traits, between individuals. For example, the spread of directly transmitted infectious diseases will be linked to network structure and this could result in social covariance in survival if infection elevates mortality. Therefore, the survival probability and capture histories of individuals in the same network are expected to change according to the histories of all individuals in that network.
Moreover, there may an additional level of clustering, with certain networks, which are in close proximity for example, affecting each other, maybe to a less, but still not negligible extent. Additionally, it is possible that network structure changes over time, with network membership, and as a result network behaviour, also changing over time. Capture-recapture models offer a possibility of modelling network structure, while accounting for changing membership over time.
A wide range of statistical social network models have been developed, especially within sociology, and are increasingly applied across a range of fields (Cranmer et al., 2017; Silk et al. 2017). However, no such modelling approach exists for capture-recapture data. Ignoring a potential network structure, and hence ignoring dependence between individuals in the same network or dependence between linked networks, may lead to biased estimates of demographic parameters of interest, such as survival probability. Further, incorporating network structure within existing capture-recapture models offers a statistical framework to understand longer-term network dynamics that is more robust to turnover in membership than many existing statistical models of networks.
The project will develop network models for capture-recapture data, motivated by a long-term data set on badgers. The European badger Meles meles is a medium-sized carnivorous mammal widely distributed across the Western Palearctic. In high density populations badgers live in territorial social groups, and individuals interact frequently with other group members and much more rarely with individuals from neighbouring groups (Weber et al. 2013 Current Biology). One such high density population in South West England has been studied using mark-recapture methods for over 40 years providing a unique long-term demographic dataset ideally suited to investigate the questions outline above (McDonald et al., 2018). Capture-recapture models (e.g. McDonald et al., 2016) and social network approaches (e.g. Weber et al., 2013, Silk et al. 2018) have been used to provide valuable insights into this population, but have yet to be combined within a single approach. Therefore, this project offers an important opportunity to gain a new understanding as to how social interactions relate to key demographic processes.
Although the project is motivated by a capture-recapture data set, the methods developed and applied will be more generally applicable to the study of network data. Given the size and complexity of the available data set, the developed methods will need to be scalable to large data sets, making them even more appealing and relevant in the study of network data.
References
Cranmer, S. J., Leifeld, P., McClurg, S. D., & Rolfe, M. (2017). Navigating the range of statistical tools for inferential network analysis. American Journal of Political Science, 61(1), 237-251
Krause, J., James, R., Franks, D. W., & Croft, D. P. (Eds.). (2015). Animal social networks. Oxford University Press, USA.
McCrea, R. S., & Morgan, B. J. (2014). Analysis of capture-recapture data. CRC Press.
McDonald, J. L., Robertson, A., & Silk, M. J. (2018). Wildlife disease ecology from the individual to the population: Insights from a long‐term study of a naturally infected European badger population. Journal of Animal Ecology, 87(1), 101-112.
Pinter-Wollman, N., Hobson, E. A., Smith, J. E., Edelman, A. J., Shizuka, D., De Silva, S., … & Fewell, J. (2013). The dynamics of animal social networks: analytical, conceptual, and theoretical advances. Behavioral Ecology, 25(2), 242-255.
Silk, M. J., Croft, D. P., Delahay, R. J., Hodgson, D. J., Weber, N., Boots, M., & McDonald, R. A. (2017). The application of statistical network models in disease research. Methods in Ecology and Evolution.
Silk, M. J., Weber, N. L., Steward, L. C., Hodgson, D. J., Boots, M., Croft, D. P., … & McDonald, R. A. (2018). Contact networks structured by sex underpin sex‐specific epidemiology of infection. Ecology letters, 21(2), 309-318.
Weber, N., Carter, S. P., Dall, S. R., Delahay, R. J., McDonald, J. L., Bearhop, S., & McDonald, R. A. (2013). Badger social networks correlate with tuberculosis infection. Current Biology, 23(20), R915-R916.
Wey, T., Blumstein, D. T., Shen, W., & Jordan, F. (2008). Social network analysis of animal behaviour: a promising tool for the study of sociality. Animal behaviour, 75(2), 333-344.
Applicants should have a good degree in statistics, mathematics, computer science, or related subjects with a strong numerical component. They should be comfortable working with data and learning new methods, determined, and interested in engaging with the practical applications of their research.
Supervisors
The supervisory team brings together multidisciplinary expertise covering statistics and ecology.
Dr Eleni Matechou and Dr Xue Wang, University of Kent
Dr Matthew Silk, University of Exeter
This is a collaborative project between the University of Kent and the Centre for Environment, Fisheries & Aquaculture Science (Cefas)
Click here to submit your application by the 8th of January.
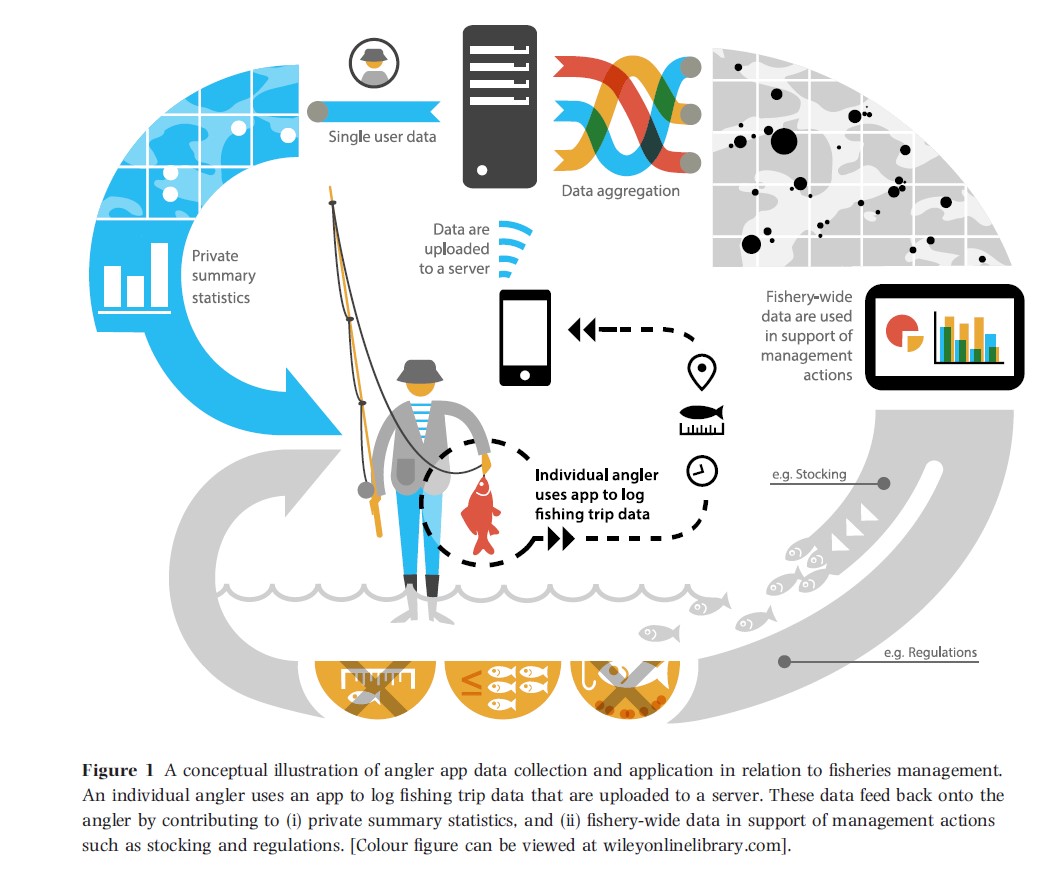
The project is at the intersection of big data, citizen-science, and sustainable fisheries. The student will provide new analytical tools for fisheries scientists to understand fish distributions, catches, effort, and angling behaviour through development of new statistical methods for the analysis of data collected by the smartphone-app Fishbrain, alongside information from current surveys.
The student will lead the development of state-of-the-art statistical methods on the timely topic of inference from large citizen-science data collected using new technologies. They will develop high-level, highly transferable statistical, programming and data skills, working with large app-derived data-sets and designed surveys using tools such as R, Python and Stan. The work will be presented and communicated to statisticians, fisheries-experts, and policy-makers at national and international conferences and meetings.
As a member of the Statistical Ecology @ Kent group and the National Centre for Statistical Ecology the student will be exposed to the latest developments in the fields of statistics and ecology. As part of the cohort of 80 Cefas PhD students they will interact with scientists and advisors from a diverse range of marine and freshwater sciences.
Through supervision and time visiting Cefas, the student will experience working in a multi-disciplinary science organisation and learn how their research fits into the wider policy context. Their work will be part of an MRF research programme at Cefas, Ball State and Danish Technical Universities, and broader fisheries advice through ICES.
The acquired knowledge and expertise in the topic of citizen-science data collected using new technologies will be of great benefit to the student in their future in academia, government, or industry. Although the models will be motivated by angling, the methods will be much more generally applicable to app-collected data on individual behaviour. The available data-sets are large and require efficient, sophisticated algorithms that fit models in reasonable time. Hence the project will equip the student with valuable skills in the growing areas of big data and data-mining.
Additional information on the project can be found here
ARIES is committed to equality & diversity, and inclusion of students of any and all backgrounds. All ARIES Universities have Athena Swan Bronze status as a minimum.
Students with high level numerical skills will be eligible for 3 months of additional stipend after the end of the 3.5 years to take advanced-level courses in branches of environmental sciences related to the project in the first 3-6 months of study.
Applicants should have a good degree in statistics, mathematics, computer science, or related subjects with a strong numerical component. They should be comfortable working with data and learning new methods, determined, and interested in engaging with the practical applications of their research.
Shortlisted applicants will be interviewed by ARIES on the 26th/27th February 2019, with shortlisting taking place at the University of Kent on the 31st of January.
Successful candidates who meet UKRI’s eligibility criteria will be awarded a NERC studentship – in 2018/19 the stipend is £14,777. In most cases, UK and EU nationals who have been resident in the UK for 3 years are eligible for a full award.
Supervisors
Dr Eleni Matechou and Dr Maria Kalli, University of Kent
David Maxwell and Dr Kieran Hyder, Centre for Environment, Fisheries & Aquaculture Science
Dr Christian Skov, National Institute of Aquatic Resources
Dr Paul Venturelli, Ball State University
The supervisory team brings together multidisciplinary expertise covering statistics, data science, recreational fisheries, app development, monitoring, and policy.
Funding Notes
The project has been shortlisted for up to 4 years, with 3.5 years minimum, of funding by the ARIES NERC Doctoral Training Partnership with a stipend of £14,777 per annum and a generous training and travel budget for attending UK-based and international conferences.

The work presents novel dynamic mixture models for the monitoring of bumblebee populations on an unprecedented geographical scale, motivated by the UK citizen science BeeWalk.
The models allow us for the first time to estimate bumblebee phenology and within-season productivity, defined as the number of individuals in each caste per colony in the population in that year, from citizen science data.
All of these parameters are estimated separately for each caste, giving a means of considerable ecological detail in examining temporal changes in the complex life cycle of a social insect in the wild. Due to the dynamic nature of the models, we are able to produce population trends for a number of UK bumblebee species using the available time-series. Via an additional simulation exercise, we show the extent to which useful information will increase if the survey continues, and expands in scale, as expected.
Bumblebees are extraordinarily important components of the ecosystem, providing pollination services of vast economic impact and functioning as indicator species for changes in climate or land use. Our results demonstrate the changes in both phenology and productivity between years and provide an invaluable tool for monitoring bumblebee populations, many of which are in decline, in the UK and around the world.

Congratulations to Anita who graduated on the 19th of July 2018 with a PhD in Statistics!
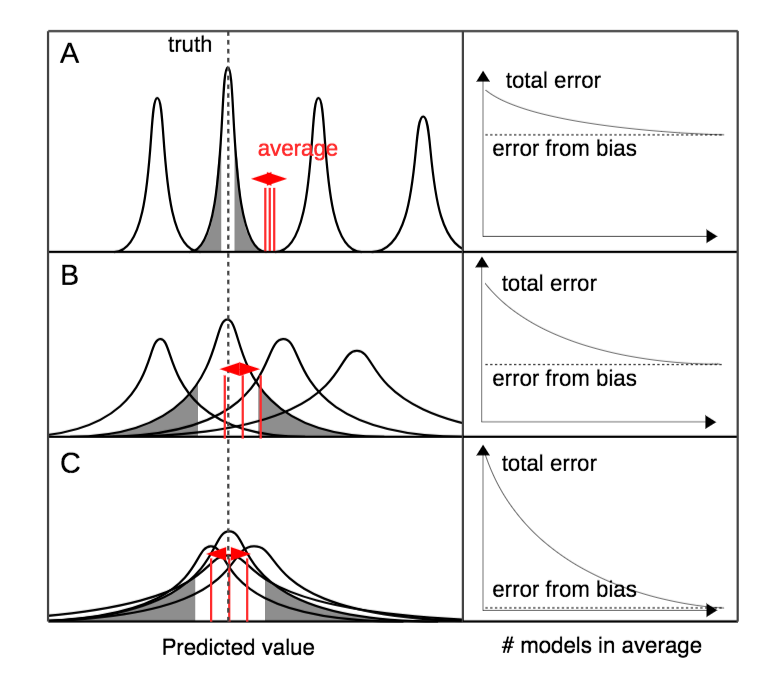
The review paper, by Dormann, C.F., Calabrese, J.M., Gurutzeta, G., Matechou, E., Bahn, V., Bartoń, K., et al. to appear in Ecological Monographs, explores different model averaging techniques in terms of ways to calculate the model weights and to combine predictions from different models. The advantages and disadvantages of model averaging are discussed and code for methods falling under three categories (Bayesian, information theoretical and tactical) is provided.
Read a blog post written by the main contributors of the paper here.

Statistical models for wildlife population assessment and conservation
Further details of the workshop and details of how to apply for a place can be found here:
https://www.kent.ac.uk/graduateschool/skills/advancedtraining.html
Please note that the deadline for applications is 1st October 2017. Successful applicants will be informed in early September.
We have 30 fully-funded places (inc. travel and accommodation) and priority is given to NERC-funded PhD students but if spaces remain we are able to offer the funded places to other PhD students and early-career researchers.
Within the environmental sector there is currently a shortage of practitioners equipped with the statistical modelling skills to carry out reliable population assessments. Consequently, environmental impact assessments (EIAs) and development mitigation projects often use population assessment protocols that are not fit-for-purpose1. The skills shortage arises because (1) recent advances in statistical models for population assessment are largely confined to the academic sector with little penetration to the end-users; and (2) although many postgraduate programmes have a statistical modelling training component, this often fails to expose PhD students to new models in the area and the potential applications these have for conservation practice2. This training programme will provide a cohort of PhD students and early career researchers/practitioners with the relevant modelling skills required for a career that involves wildlife population assessment for conservation.
Proposed programme of the course
The workshop will focus on ecological questions that arise in conservation practice and use real case study data. Training will include individual-based models, such as capture-recapture, but will also embrace scenarios more frequently used in EIA, such as batch-marked, presence/absence, site occupancy and counts. Applications will include newts, butterflies, birds, bees, beetles, ibex and bats. Each module will be accompanied by a practical computer session using R and each module builds on the last so that delegates build a portfolio of statistical skills.
Training outcomes: By the end of the course, attendees will be able to:
Eleni gave a seminar on the 15th of March 2017 as part of the Ecology, Evolution and Environment seminar series at the Department of Animal and Plant Sciences, University of Sheffield.
Title: Modelling phenology for marked and unmarked populations
Abstract: In this seminar I will discuss fairly recent models for capture-recapture and for count data that enable us to estimate, among other things, phenology of wildlife populations. The methods explored will include classical as well as Bayesian parametric and non-parametric approaches. They will be demonstrated using data on breeding great crested newts, migrating reed warblers, bivoltine butterfly species , bumblebees from the citizen science scheme BeeWalk as well as data on anglers in Norway.
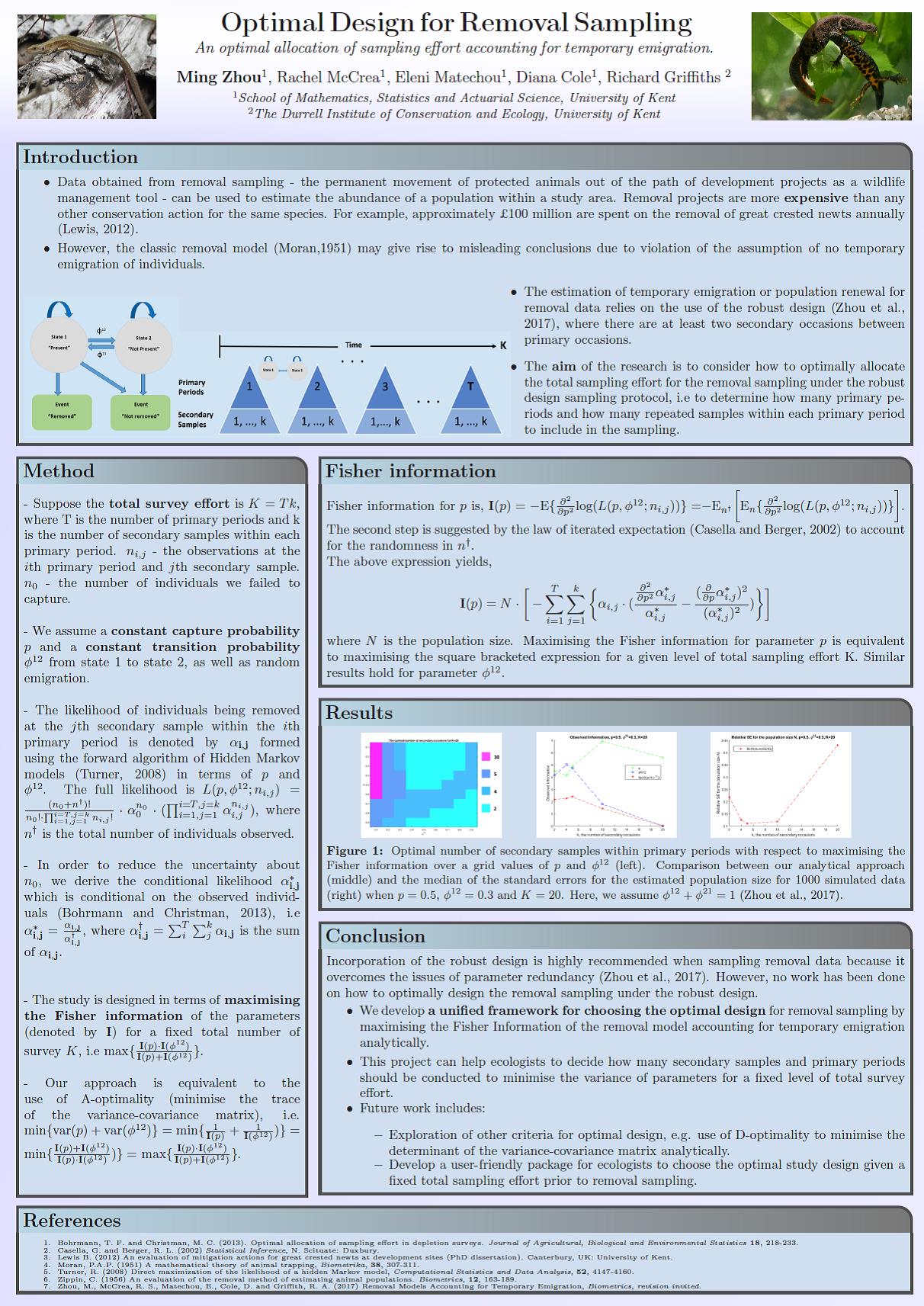
Ming presented a poster titled “Optimal Design for Removal Sampling” at the postgraduate research festival that took place at the University of Kent.
She presented her work in which she investigates removal models accounting for temporary emigration analytically and examines how to optimally allocate a fixed level of total sampling effort in terms of maximising the Fisher information.

Byron has been awarded a 24-month Leverhulme Emeritus Fellowship to enable him to conduct research on “Environmental modelling for moths and butterflies”.
Well done Byron!