Watch the short documentary highlighting the work of the SRT, which also includes members of SE@K, such as Bruno, Daniel and Eleni.

STEM for Britain 2023
Alex presented his work on modelling metabarcoding data at the 2023 STEM for Britain Competition
His poster described the novel modelling framework, introduced in the corresponding paper, and showed results from a data set on ingested DNA from a survey in China.

SE@K Thursday lunches – Summer 2023
For the last term of the 22/23 academic year, we have an exciting plan for SE@K and guest talks
- 11th of May Session : eDNA – Alex/Eleni; Cake : Eleni
- 18th of May Session : Occupancy data – Ardian Ardiantiono; Cake : James
- 25th of May Session : BeeWalk modelling – Fabian; Cake : Fabian
- 1st of June Session : Elephant killings – Kennedy Leneuiyia; Cake : Milly
- 8th of June Session : Extinction Models – Dave Roberts; Cake : Bruno
- 15th of June Session : Binary time series – Takis (TBC); Cake : Daniel
- 22nd of June Session : PhD student NCSE presentations; Cake : Diana

Bruno and Eleni visited Stockholm
The visit was part of their Swedish research council funded project on modelling overcoverage in population registers, in collaboration with Dr Eleonora Mussino.
The first paper from this project is already under review, using multiple systems estimation to estimate the number of immigrants who are present but have not been observed in any register each year.
They are currently developing capture-recapture-recovery models with temporary emigration as an alternative approach for estimating the probability that an individual is there given that they weren’t observed.
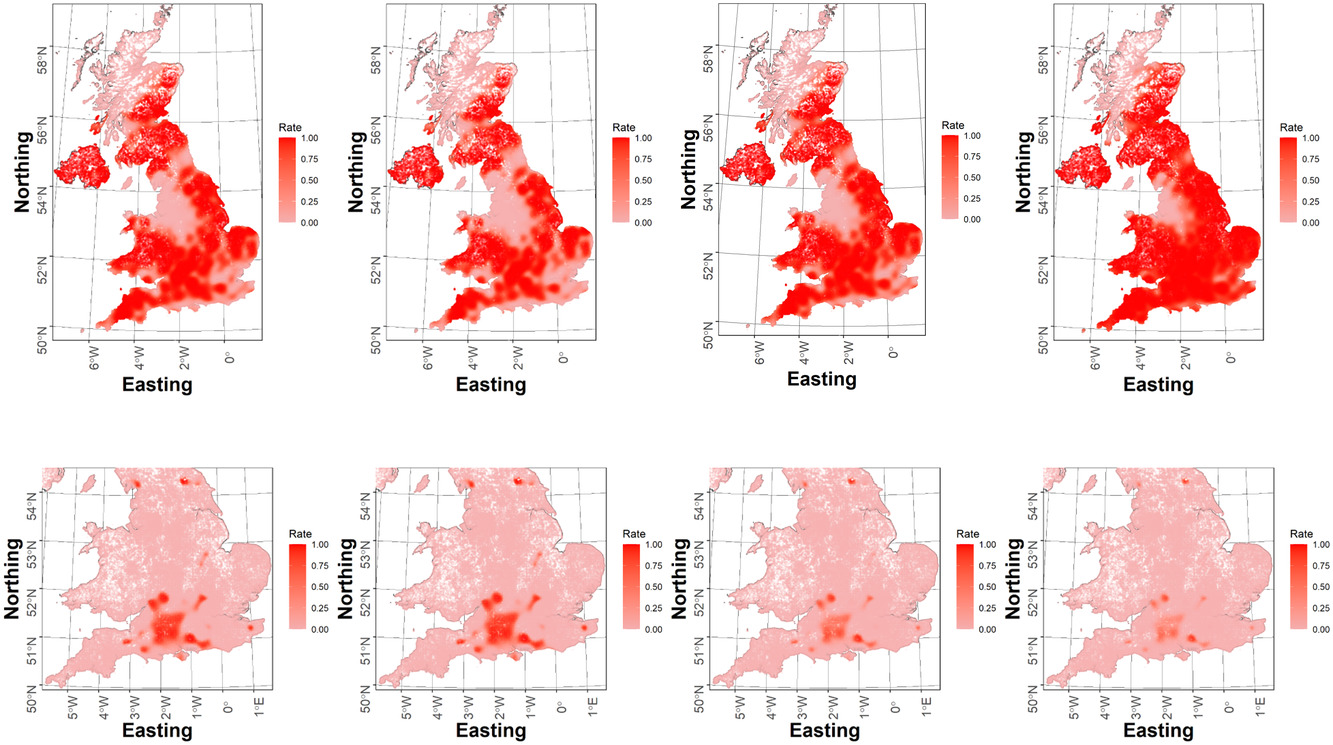
New Biometrics paper
A new modelling framework for fast Bayesian inference in large occupancy data sets by Alex, Emily, Eleni and Byron just published in Biometrics.
The model accounts for spatio-temporal autocorrelation and gives robust inference on species presence, as evidence by the simulation results in the paper and the two case studies on one common and widespread and one rare species, using records from the Butterflies for the New Millennium database, producing occupancy indices spanning 45 years.

Spring term 2023 SE@K Thursday lunches
Spring term starts with the second half of Python training by Lena :-). Plan for the term in terms of sessions (and cake) below!
- 19th of January Session : Python training II – Lena; Cake : Lena & Daniel
- 26th of January Session : Daniel’s grant; Cake : Diana
- 2nd of February Session : HMM paper – Diana; Cake : Eleni
- 9th of February Session : HMM paper – Daniel; Cake : Oscar
- 16th of February Session : HMM paper – Takis (guest lecture); Cake : Alex
- 23rd of February Session : Variable selection/Hypothesis testing – Alex; Cake : James
- 2nd of March Session : HMM – Fred (guest lecture); Cake : Fabian
- 9th of March Session : Parameter redundancy – Daniel’s take; Cake : Tommy
- 16th of March Session Swedish register data – Bruno&Eleni ; Cake : Milly
- 23th of March Session : Swedish register data – Bruno&Eleni; Cake : Daniel
- 30th of March Session : Spatio-temporal models – Oscar; Cake : Diana
- 6th of April Session : eDNA data – Alex; Cake : Eleni

Se@k weekly lunches
The SE@K group are meeting weekly for research lunch and socialising 🙂
Autumn term
First week was about introductions and a tasty lunch at Dolce Vita!
Second week was about staff talking about their research interests, with Diana talking to the group about parameter redundancy.
Third, fourth & fifth week were about research students talking about their research, with Tommy using randomised response techniques to estimate the proportion of people in the group who like/liked their supervisor (!), Milly talking about distance sampling, Ioannis talking about Bayesian non-parametrics and ABC and Lena talking about predator-prey models!
The term went on with learning about Gaussian processes from Alex, about PDEs from Eduard and about Python from Lena!






Eleni visited Lille for 3i workshop on Biodiversity and its Conservation
The one-day workshop, organised by the group in Lille, consisted of a series of talks by researchers from the Universities of Lille, Ghent and Kent.
Eleni gave talks on her work on modelling BeeWalk data and on modelling eDNA data.

New paper in Environmetrics by Alex
A vector of point processes for modeling interactions between and within species using capture-recapture data
Alex Diana, Eleni Matechou, Jim E. Griffin, Yadvendradev Jhala, Qamar Qureshi
Abstract
Capture-recapture (CR) data and corresponding models have been used extensively to estimate the size of wildlife populations when detection probability is less than 1. When the locations of traps or cameras used to capture or detect individuals are known, spatially-explicit CR models are used to infer the spatial pattern of the individual locations and population density. Individual locations, referred to as activity centers (ACs), are defined as the locations around which the individuals move. These ACs are typically assumed to be independent, and their spatial pattern is modeled using homogeneous Poisson processes. However, this assumption is often unrealistic, since individuals can interact with each other, either within a species or between different species. In this article, we consider a vector of point processes from the general class of interaction point processes and develop a model for CR data that can account for interactions, in particular repulsions, between and within multiple species. Interaction point processes present a challenge from an inferential perspective because of the intractability of the normalizing constant of the likelihood function, and hence standard Markov chain Monte Carlo procedures to perform Bayesian inference cannot be applied. Therefore, we adopt an inference procedure based on the Monte Carlo Metropolis Hastings algorithm, which scales well when modeling more than one species. Finally, we adopt an inference method for jointly sampling the latent ACs and the population size based on birth and death processes. This approach also allows us to adaptively tune the proposal distribution of new points, which leads to better mixing especially in the case of non-uniformly distributed traps. We apply the model to a CR data-set on leopards and tigers collected at the Corbett Tiger Reserve in India. Our findings suggest that between species repulsion is stronger than within species, while tiger population density is higher than leopard population density at the park.