The model has been developed for single species qPCR data. The data are the number of positive qPCRs (eDNA score) for each water sample collected at surveyed sites.
The model allows us to estimate the probability of species presence at each surveyed site, while accounting for the probabilities of a false positive and false negative error at stage 1 (field) and stage 2 (lab).
The model parameters are

A schematic representation of the model is shown below
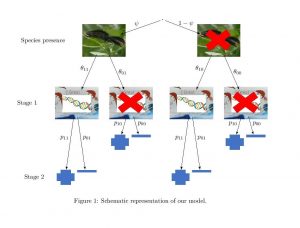
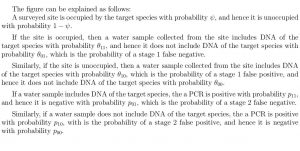
The model is fitted within a Bayesian framework and any of the model parameters can be functions of covariates. The implemented algorithm performs Bayesian variable selection and the output includes posterior summaries of all parameters as well as posterior probabilities of inclusion (see examples and Griffin, J. E., Matechou, E. Buxton, A. S., Bormpoudakis, D. and Griffiths, R. A., (2020) Modelling environmental DNA data; Bayesian variable selection accounting for false positive and false negative errors, Journal of the Royal Statistical Society: Series C (Applied Statistics) for a more detailed description of how to interpret the output).
