Working alongside Prof. Michelle Garrett at the School of Biosciences, I will be laying the groundwork for a very different way to tackle difficult diseases thanks to funding from the Rosetrees Trust. We will look particularly at a protein which is involved in 30% of cancers, but whose behaviour cannot be altered by any current drugs. This funding will also support a PhD student. You can find out more about this “Postgraduate Scholarship in Discovery of Therapeutic Sequence-Defined Polymers” opportunity here.
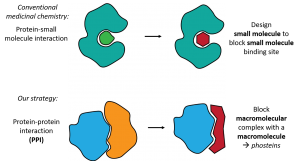
There are critical steps in almost every disease process involving multiple large molecules (macromolecules) called proteins, coming together in the cell. These protein-protein interactions (PPIs) are central to diseases including cancer, Alzheimer’s, HIV and cystic fibrosis, yet they are exceedingly challenging to block using conventional approaches. This is because classical drug discovery relies on small molecules sitting within well-defined pockets inside proteins. Conversely, PPIs involve large, comparatively featureless surfaces of macromolecular proteins, which cannot be effectively blocked by a small molecule. We propose a new approach: to block a macromolecular interaction between proteins, we should use synthetic macromolecules.
This grant funds the groundwork for an entirely new approach to the drugging of PPIs using synthetic macromolecules. I have developed a novel class of macromolecules which, like proteins, are made by linking smaller units in different orders to make macromolecules with specific sequences. Like proteins, the sequence determines 3D structure, and interaction with other molecules. Due to this similarity to proteins, and because each unit contains a phosphate group, we call these novel macromolecules ‘phosteins.’ Phosteins are ideal for blockade of PPIs because their size lets them bind the entire PPI surface, and their synthetic nature means they can be tailored at will.
But, out of the many millions of possible sequences, how will we find a phostein that can block a PPI? To do so, we need to use an ‘evolutionary’ method which works by selection-of-the-fittest. This involves adapting instruments capable of cells to sort a massive library of sequences into those which bind and those which don’t. We will use this automated selection to discover phosteins which bind RAS proteins – which frequently drive cancers, but are currently intractable drug targets – if we can stop RAS associating with its partner proteins, it will be a major breakthrough. Moreover, in the future, the approach could be used to address PPIs in all kinds of other diseases.
We’re looking for strong applicants with a background in chemistry or biochemistry and an enthusiasm for working across subject boundaries. Further details on the studentship can be found at http://www.jobs.ac.uk/job/BIF213/postgraduate-scholarship-in-discover-of-therapeutic-sequence-defined-polymers/
Dr Chris Serpell – Lecturer in Chemistry

