Biology requires the seamless working together of genes and proteins.
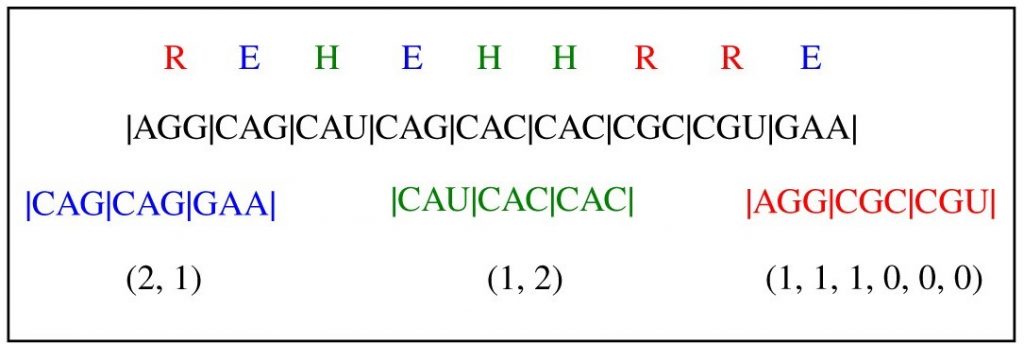
Genes encode information used for the synthesis of proteins which provide the chemical activities essential for life. Due to the way genes encode this information, the same protein can be made by many different gene sequences. Such sequences are not completely equivalent though, as they typically have different efficiencies for information storage.
How gene sequences evolve to produce the right protein in the right amount is a hotly researched question. Gene sequences are shaped by many different forces, including the need to avoid internal base pairings, a requirement to bind tightly to the decoding machinery, and a need interact with other control molecules. Existing studies have often focussed on just one to these forces, rather looking at the system as a whole.
Researchers in the Schools of Computing and Biosciences at Kent has taken the approach of looking at the system as a whole. Their work takes inspiration from Austrian physicist Ludwig Boltzman who in the 1860s was interested in the speed of atoms in gases. This system too is shaped by a large number of individual forces which makes the overall behaviour complicated to model and understand. However, Boltzman used statistical methods to characterise the overall effect without needing to look at each individual force in detail. The Kent researchers have used a similar approach to study the evolution of gene sequences and discovered that the balance of forces is variable in fungi and bacteria, two groups of microbes with strong industrial and medical applications.
Image: more than one gene sequence can code for the same protein