Professor Darren Griffin and Dr Becky O’Connor, in collaboration with Dr Denis Larkin at the Royal Veterinary College, London have recently been awarded a 3-year BBSRC grant for a study entitled “Rapid reconstruction of reference chromosome-level mammalian genome assemblies and insight into the mechanisms of gross genomic rearrangement”
We live in an era in which the genomes of new species are being sequenced all the time. The most modern ways to sequence DNA have many advantages over older approaches (the prominent one being a vastly reduced cost) but a problem that arises each time the genome of a new species is sequenced is that assigning large blocks of sequence to an overall genomic “map” can be problematic and/or very expensive. It’s a little like finding your location on Google Maps but not being able to “zoom out” to establish where that position is in relation to the whole country. In essence, the aim of this project is to rectify this problem at one fifth of the current cost. Using our experience with birds we have developed a high-throughput approach and the tools for assigning the sequences to their proper positions in chromosomes. This involves our own adaptations to a technique called “FISH” that can take the data from sequenced genomes and visualize directly blocks of DNA sequence as they appear in their rightful place in the genome.
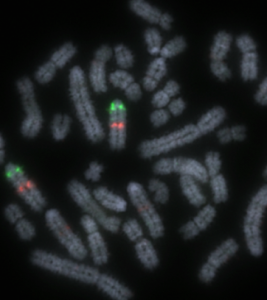
Sequences of the pig genome visualised on chromosomes using fluorescent probes (FISH)
In this study, we will focus our attention on 25 newly sequenced mammal species. More importantly however we will provide the means through which this can be achieved for any of the 5,000 living mammalian species. Mammals are important to our lives in that many are models for human disease and development and are critical to agriculture (both meat and milk). Others are threatened or endangered and, with impending global warming, molecular tools for the study of their ecology and conservation are essential. Our combined efforts have also developed computer-based browser methods to compare the overall structure of one genome with another, directly visualizing the similarities and differences between the genomes of several animals at a time, something we can share widely amongst the scientific community and general public via the world wide web. The differences between mammalian genomes arose through changes that happened during evolution. One of the main aims of this project is to find out how this occurred and what are implications of these changes. We have a number of ideas such as we think there may be different “signatures” that classify why blocks of genes tend to stay together during evolution. Armed with this information, we fully intend to take it out into the world. The devices that we will develop can be adapted for the screening of individual animals for genomic rearrangements that may cause e.g. breeding problems. Moreover, the resources we will develop provide a source for public information and student learning through a dedicated, outwardly-facing web site. We have received overwhelming support from numerous laboratories all over the world who are interested in using the resources that we will develop to ask biological questions of their own. For this reason, we feel that this project will help us understand evolution in mammals and contribute to establishing the UK as a central international hub of mammalian genomics.